PROBLEM:
Identify the feasibility of using machine learning approaches for medical image auto-segmentation (auto-contour) of the bladder from DICOM image set and manual contour set (10 anonymized patients) provided by our customer.
PARTICIPANTS:
Nghia.TD@3si.vn, Nam.VH@3si.vn
DURATION: 1 week
DICOM Data reception from customer: May 25, 2017
APPROACHES (May 25 – May 28):
We used the following:
Hardware acceleration platform
- Intel-based CPU with NVIDIA GPU for multicore and parallel computing acceleration.
Software platform
We plan to try the follow 2 systematic approaches using some proven Deep Learning technique with Convolutional Neural Networks (CNN)
1. Deep Learning with Neural Network:
A number of different approaches have been used to try to address this goal.
1.1. Build a system to run Medical Image Segmentation using Alexnet neuron network (converted to Fully Convolutional Network (FCN) model) on DIGITS (the NVIDIA Deep Learning GPU Training System [1]).
1.2. Using pre-trained CNN, Microsoft Cognitive Toolki, and Azure GPU VMs
1.3. Using 3D U-Net CNN
1.4. Using SegNet
1.5. Using 3D CNN and level sets
2. Usage of existing service from our partnership with IBM (IBM GPSG & IBM Vietnam) with Watson Health.
PROGRESS (May 29 – Present):
- After contacting IBM to share the PROBLEM above that need total solution in Watson Health. So far, it is still in internal searches within IBM networks for a similar example of implementation in similar kind of medical image segmentation.
- Deep learning with DIGITS system:
Platform preparation: DONE
Hardware
- Intel Dual Xeon E5v3 2683 (Haswell E/EP):56 cores @2GHz
- NVIDIA GTX 780 Ti 3GB x 4 (2880×4 = 11520 CUDA cores) – KEPLER Architecture
- 96GB DDR4 RAM
- SATA 512GB + 2TB HDD Raid 0
Software
- Ubuntu 16.04 LTS
- Full set of DIGITS (NVIDIA Deep Learning GPU Training System) with Caffe, Caffe2, modified 3D-Caffe, Tensorflow, Torch 7, OpenCV 2 & OpenCV 3, Python 2.7 & 3.5, MXnet (and their corresponding docker containers images)
- MITK toolset (super-build) and ImageJ, Aeskulap for dicom viewer and data preparation, examination and info extraction.
Concept
In theory, we (1) take some data, (2) train a model on that data, and (3) use the trained model to make predictions on new data.
Implementation
Left Ventricle Open Online Segmentation Challenge
We refer to the system set-up and running on an old online computer science challenging. [2]
The model was created pre-trained by fcn_alexnet.caffemodel and python layer of Dice Coefficient to obtain accuracy (in the provided sample) uup to 63% and can be further optimization using Dice Coefficient as a Loss layer, moving to FCN8s model (fcn8s-heavy-pascal.caffemodel), etc. The current Kepler GPU with only 3GB VRAM each so that it tthrowsOut of Memory error on finer fcn8s models.
UPDATED 06/15:
The system is upgraded to replace with 1 Nvidia GTX1080 GB and the fcn8s was successfully deploy and trained on the current database. This brought the dice (val) up from 62% to 82% in the new deep learning model, A significant improvement that brings the accuracy and smoothness of the auto-contour (Figure 1.b).
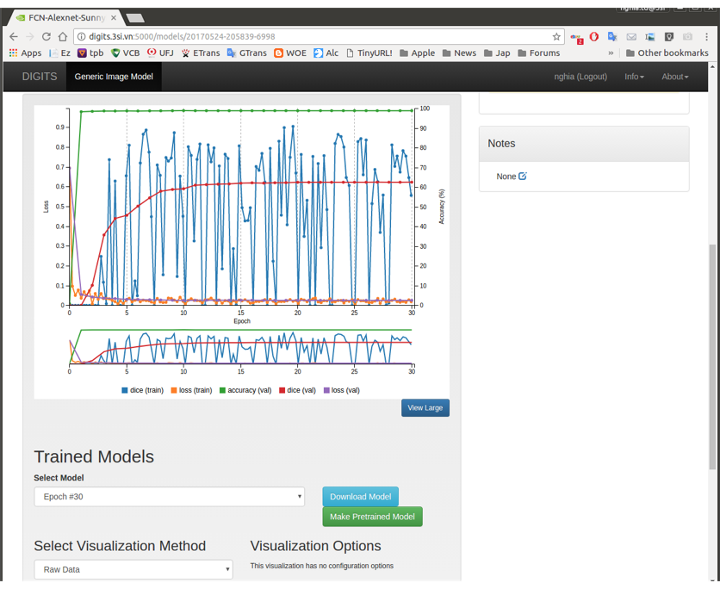
Figures 1.a: The screenshot of actual FCN model creation for LV Challenge on 3S system with GTX 780 Ti 3Gb
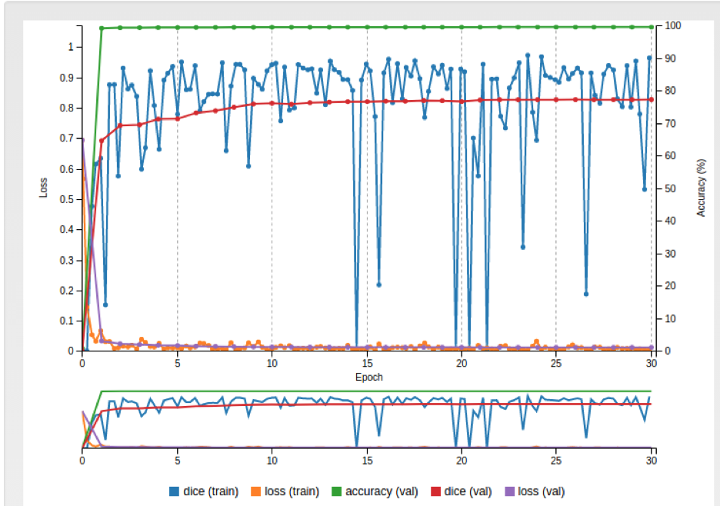
Figures 1.b: The screenshot of actual FCN8S model creation for LV Challenge on 3S system with GPU GTX 1080 8Gb
After training the model, we run some sample tests to confirm the model is actually working smoothly on our system. With samples of Left Ventricle system, the system is able to recognize, auto contour and segmentize the target area.
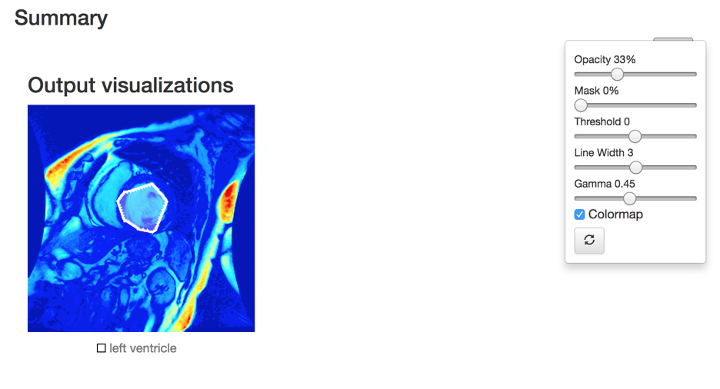
Figure 2.a: the screenshot of actual model inference result test on LV sample (Output visualization in colormap).
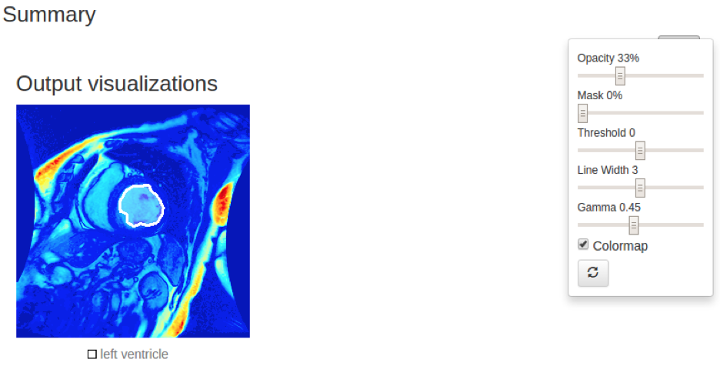
Figure 2.b: the screenshot of actual model fcn8s inference result test on LV sample (Output visualization in colormap).
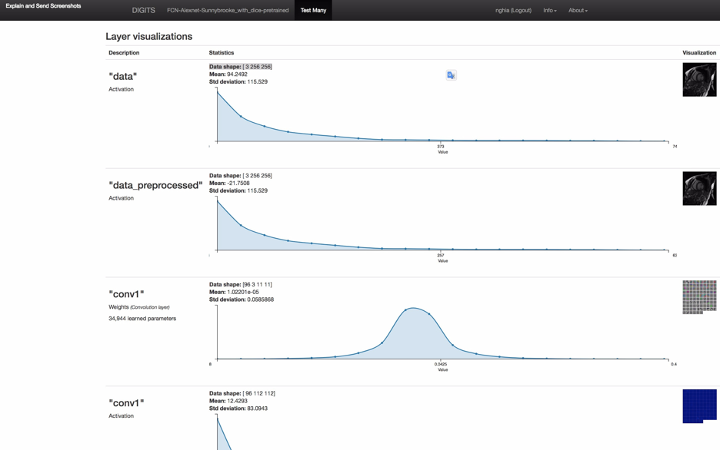
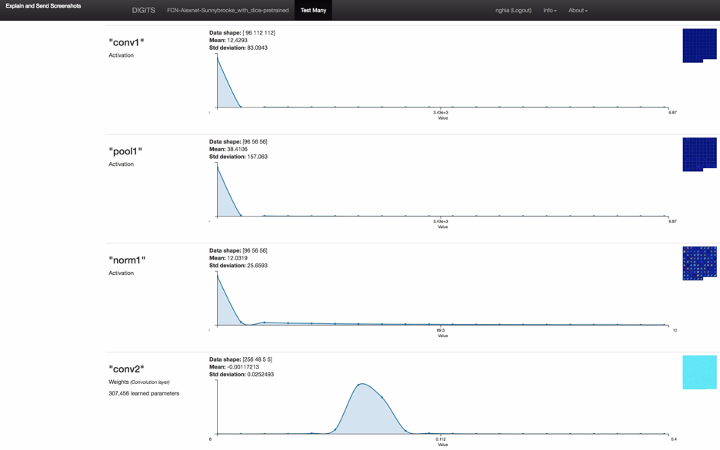
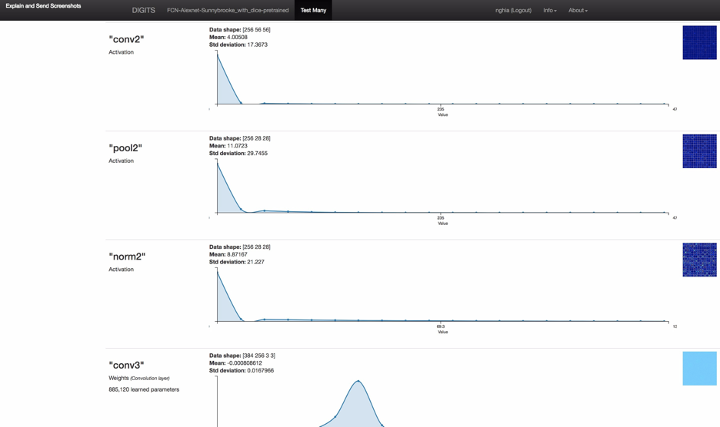

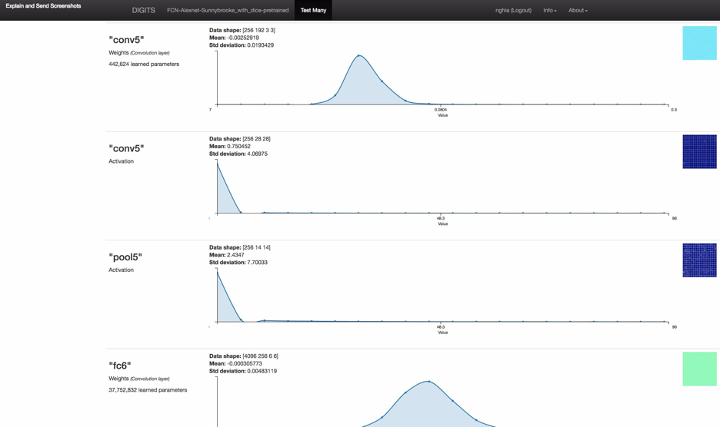
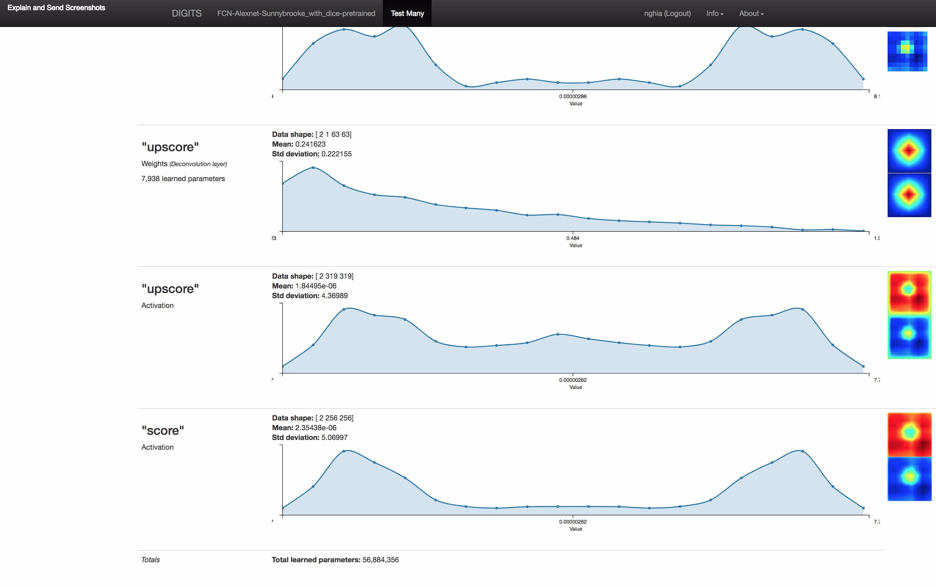 Figure 3: the screenshot of full model inference result including neuron network layer visualizations
Figure 3: the screenshot of full model inference result including neuron network layer visualizations
The first of all is to do the data pre-processing and convert provided images set into the valid dataset of training, evaluation, and test. It takes a while in order to comprehend the dicom images of slices and the DICON Structure Set (RS). On my Ubuntu 16.04 LTS system, not many free tools support reading normal DICOM and it is even less tools to be able to read and display DICOM RS. Finally, I have to compile an open-source Medical Imaging Interaction Toolkit (MITK) [4] myself from its source-code and manage to read a completed dicom dataset.
To run our PROBLEM against the above pretrain model setup we have to have (1) take the data:
- DICOM CT of the bladder area (DONE)
- Extract the contour into a matrix of coordinate point inner and outer contour of the bladder (NOT DONE). The matrix format for each slice should be:
IM-0001-0020-icontour-manual.txt 120.00 93.00
120.50 93.00
121.00 92.50
121.50 92.50
122.00 92.50
122.50 92.00
123.00 92.00
123.50 92.00
124.00 91.50
124.50 91.50
125.00 91.50
…………
IM-0001-0020-ocontour-manual.txt 111.50 99.00
112.00 98.50
112.50 98.00
113.00 97.50
113.50 97.00
113.50 96.50
114.00 96.00
114.50 96.00
115.00 95.50
115.50 95.00
116.00 94.50
…………
(2) Train a model on that data: TODO
(3) Use the trained model to make predictions on new data: TODO
3. Deep learning with pre-trained CNN, Microsoft Cognitive Toolkit and Azure GPU VMs
To achieve this, we used the following:
Software
- A pre-trained CNN as the image featurizer. This 152-layer ResNet [7] model is implemented on the Microsoft Cognitive Toolkit deep learning framework (CNTK) [5] and trained using the ImageNet [6] dataset.
- Azure Virtual Machines (VMs) [9] with GPU acceleration.The Bladder images are in DICOM format and consist of a group of horizontal slices for each patient.
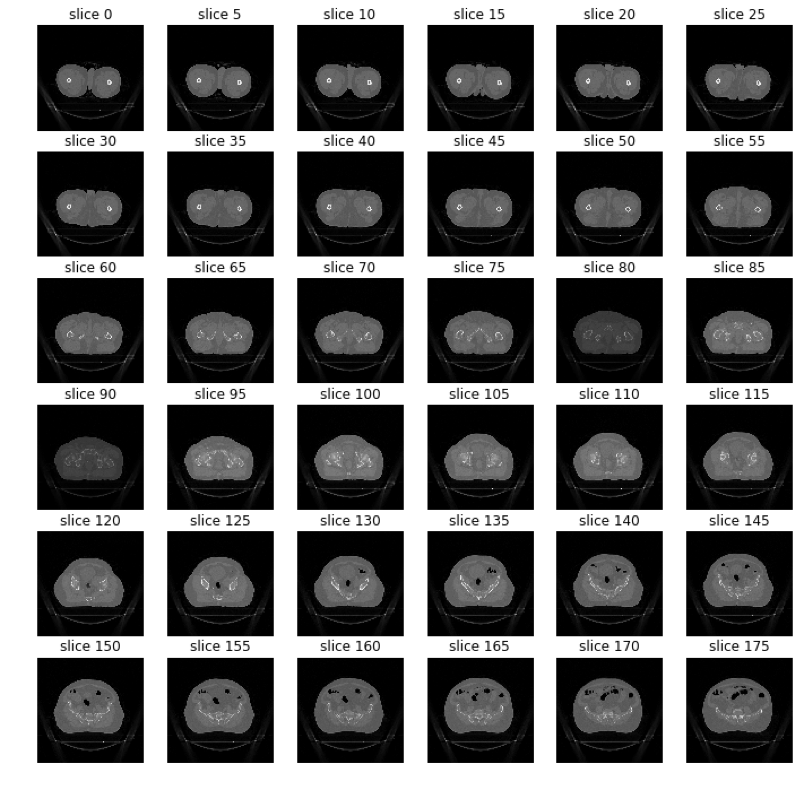
Figure 4: The slices of the bladder of a patient.
Bladder Segmentation with Cognitive Toolkit
Many deep learning applications use pre-trained models as a basis and apply trained models to a new domain, in a technique called transfer learning [8].
We use transfer learning with a pre-trained CNN on ImageNet as a featurizer to generate features from the Bladder dataset.
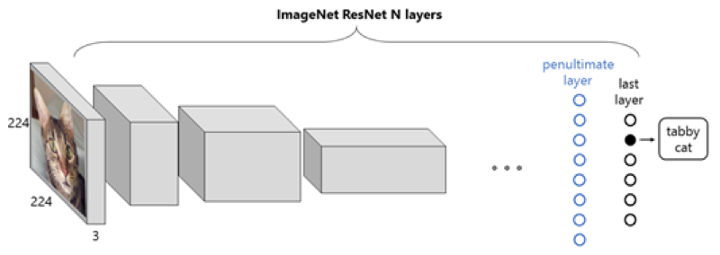
Figure 5: Representation of a ResNet CNN with an image from ImageNet. The input is an RGB image of a cat, the output is a probability vector, whose maximum corresponds to the label “tabby cat”.
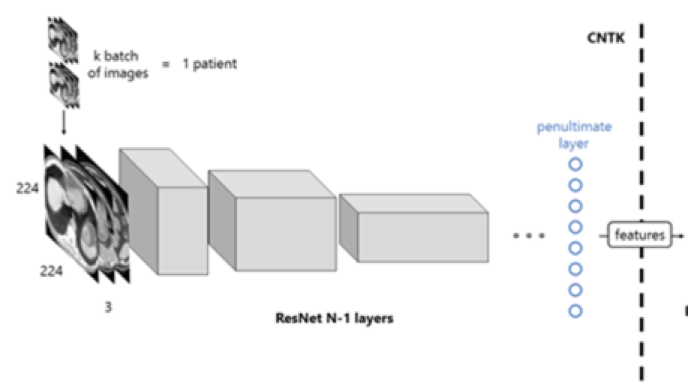
Figure 6: Workflow of the proposed solution. The images of a patient scan are fed to the network in batches, which, after a forward propagation, are transformed into features. This process is computed with the Microsoft Cognitive Toolkit.
Train a model on that data: TODO
Use the trained model to make predictions on new data: TODO
4. Deep learning with 3D U-Net CNN
Software
- python 2.7, CUDA 8.0, cudnn 5.1, h5py (2.6.0), SimpleITK (0.10.0), numpy (1.11.3), nvidia-ml-py (7.352.0), matplotlib (2.0.0), scikit-image (0.12.3), scipy (0.18.1), pyparsing (2.1.4), pytorch (0.1.10+ac9245a)
Implementation
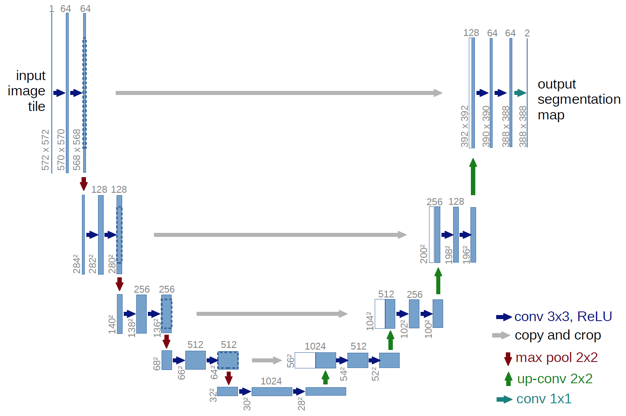
Figure 7: U-Net: Convolutional Networks for Biomedical Image Segmentation
Use the trained model to make predictions on new data: TODO
Train a Bladder data: TODO
5. Deep learning with SegNet
We refer to the Lung Image Segmentation with CNN Conference Paper [12]
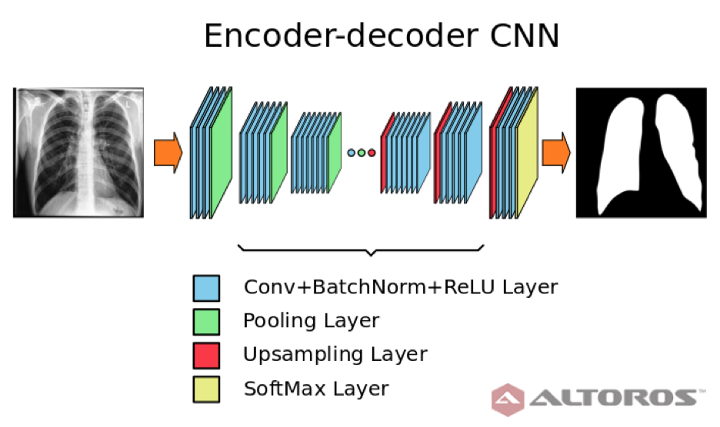
Figure 8: Simplified scheme of encoder – decoder neural network architecture
The network had a typical deep architecture with the following key elements:
- 26 convolutional layers
- 25 batch normalization layers
- 25 ReLU layers
- 5 upsampling layers
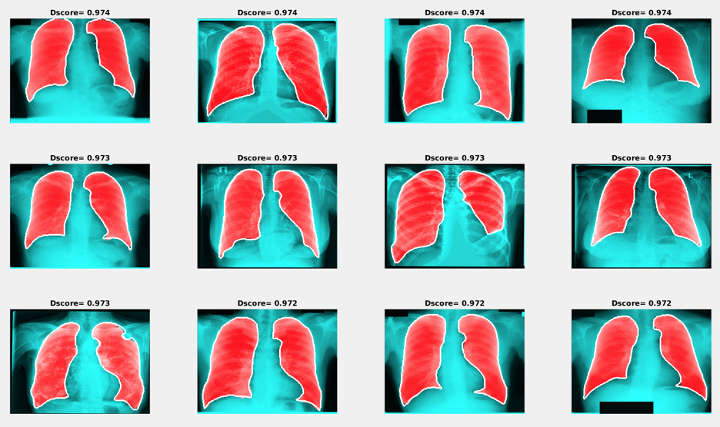
Figure 9: Examples of segmentation results with the maximum Dice score
Train a Bladder data: TODO
Use the trained model to make predictions on new data: TODO
6. Deep learning with 3D CNN and level sets
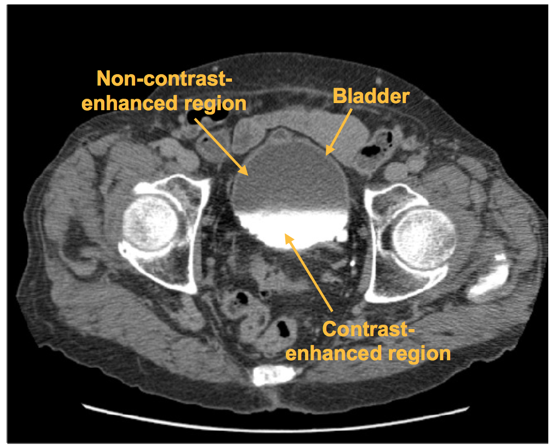
A CNN was trained to distinguish between regions of interest (ROI) that are inside and outside of the bladder. The CNN outputs the likelihood that an input ROI is inside the bladder, which is used to form the bladder likelihood map. The map is used to generate the initial contour for level-set based bladder segmentation.
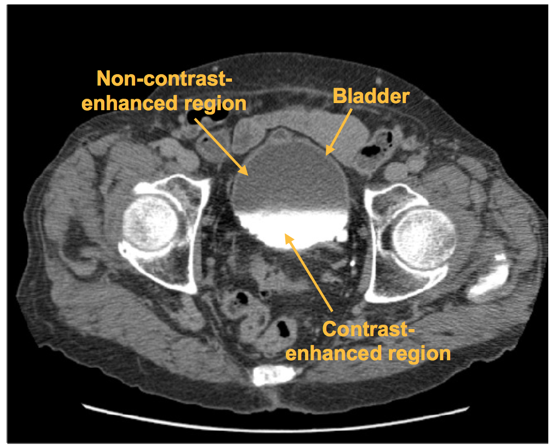
Figure 10: An axial slice of a CTU scan in which the bladder is partially filled with intravenous (IV) contrast material. Two distinct regions, the non-contrast-enhanced region, and the contrast-enhanced region, is shown. A malignant lesion is present in the contrast-enhanced region of the bladder.
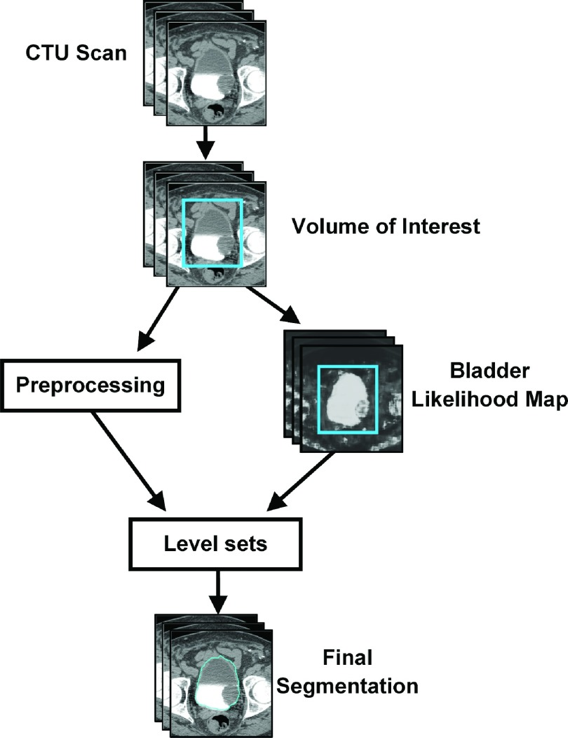
Figure 11: Flowchart of the template-based segmentation method

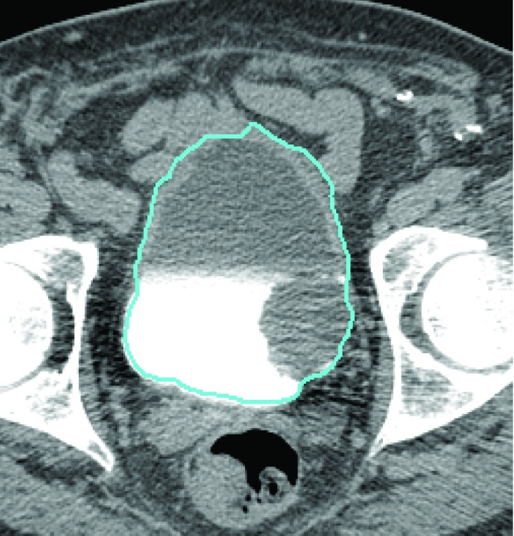
Figure 12: Block diagram of the CNN architecture
Figure 13: Bladder segmentation of the CTU slice using the CNN bladder likelihood map with level set
Implement an algorithm from a scientific paper: TODO
Use the trained model to make predictions on new data: TODO
ACTION ITEMS
- Further study on related REFERENCE researches, especially spending more time to understand prostate segmentation work from CUMED research team [4] and Kaggle Data Science Bowl 2017 [13]. Consider some other fine-tuning and experiment in computer vision, auto-contouring and image segmentation using different Deep Learning neural network adapting into DIGITS.
- Consider data pre-processing by traditional auto-segmentation/contour [11] as input for the supervised training set.
- Share further progress result to customer and decide further step of involvement into the opportunity
NEXT WEEK’S AGENDA
- Having skype conference with customer for advice on the above approaches and also exchange information on his current ML approach for Prowess system.
- Extraction of contour data into readable format by DIGITS system. Try to complete training dataset for fcn alexnet pretrained model in the LV above.
- Improve model on either upgrade added GPU to hardware platform (Pascal architecture 1080 8GB or 1080 Ti 11GB in order to run bigger / better / finer FCN model on image segmentation).
- Invite customer to 3S office for ~ 1 hour experience sharing on Health Record and images and other terminology of an OIS.
REFERENCES
[1] NVIDIA DIGITS, https://github.com/NVIDIA/DIGITS
[3] The Medical Imaging Interaction Toolkit (MITK)
[4] Volumetric ConvNets with Mixed Residual Connections for Automated Prostate Segmentation from 3D MR Images by Lequan Yu, Xin Yang, Hao Chen, Jing Qin, Pheng-Ann Heng (Department of Computer Science and Engineering, The Chinese University of Hong Kong, Centre for Smart Health, School of Nursing, The Hong Kong Polytechnic University; Guangdong Provincial Key Laboratory of Computer Vision and Virtual Reality Technology; Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, China) >>> Current leader of the Promise12 Grand Challenge namely CUMED (submission on 14/08/2016).
[5] Microsoft Cognitive Toolkit (CNTK)
[6] ImageNet
[7] Deep Residual Learning for Image Recognition
[8] Yosinski et al., 2014; Donahue et al., 2014; Oquab et al., 2014; Esteva et al., 2017
[10] Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets Kenny H. Cha, Lubomir Hadjiiski, Ravi K. Samala, Heang-Ping Chan, Elaine M. Caoili, and Richard H. Cohan Department of Radiology, The University of Michigan, Ann Arbor, Michigan
[11] Automatic segmentation of bladder in CT images by Feng SHI, Jie YANG, Yue-min ZHU ( Institute of Image Processing and Pattern Recognition, Shanghai Jiao Tong University, Shanghai 200240, China) (26/12/2008).
[12] Lung Image Segmentation Using Deep Learning Methods and Convolutional Neural Networks
Promise12 MICCAI Grand Challenge: Prostate MR Image Segmentation 2012
Retina blood vessel segmentation with a convolution neural network (U-net)